Research ArticleABSTRACT
Sunlight is the most abundant energy source on this planet. However, the ability to convert sunlight into biological energy in the form of adenosine-5′-triphosphate (ATP) is thought to be limited to chlorophyll-containing chloroplasts in photosynthetic organisms. Here we show that mammalian mitochondria can also capture light and synthesize ATP when mixed with a light-capturing metabolite of chlorophyll. The same metabolite fed to the worm Caenorhabditis elegans leads to increase in ATP synthesis upon light exposure, along with an increase in life span. We further demonstrate the same potential to convert light into energy exists in mammals, as chlorophyll metabolites accumulate in mice, rats and swine when fed a chlorophyll-rich diet. Results suggest chlorophyll type molecules modulate mitochondrial ATP by catalyzing the reduction of coenzyme Q, a slow step in mitochondrial ATP synthesis. We propose that through consumption of plant chlorophyll pigments, animals, too, are able to derive energy directly from sunlight.
INTRODUCTION
Determining how organisms obtain energy from the environment is fundamental to our understanding of life. In nearly all organisms, energy is stored and transported as adenosine-5′-triphosphate (ATP). In animals, the vast majority of ATP is synthesized in the mitochondria through respiration, a catabolic process. However, plants have co-evolved endosymbiotically to produce chloroplasts, which synthesize light-absorbing chlorophyll molecules that can capture light to use as energy for ATP synthesis. Many animals consume this light-absorbing chlorophyll through their diet. Inside the body, chlorophyll is converted into a variety of metabolites (Ferruzzi and Blakeslee, 2007; Ma and Dolphin, 1999) that retain the ability to absorb light in the visible spectrum at wavelengths that can penetrate into animal tissues. We sought to elucidate the consequences of light absorption by these potential dietary metabolites. We show that dietary metabolites of chlorophyll can enter the circulation, are present in tissues, and can be enriched in the mitochondria. When incubated with a light-capturing metabolite of chlorophyll, isolated mammalian mitochondria and animal-derived tissues, have higher concentrations of ATP when exposed to light, compared with animal tissues not mixed with the metabolite. We demonstrate that the same metabolite increases ATP concentrations, and extends the median life span of Caenorhabditis elegans, upon light exposure; supporting the hypothesis that photonic energy capture through dietary-derived metabolites may be an important means of energy regulation in animals. The presented data are consistent with the hypothesis that metabolites of dietary chlorophyll modulate mitochondrial ATP stores by catalyzing the reduction of coenzyme Q. These findings have implications for our understanding of aging, normal cell function and life on earth.
RESULTS
Light-driven ATP synthesis in isolated mammalian mitochondria
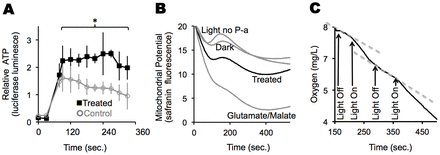
To demonstrate that dietary chlorophyll metabolites can modulate ATP levels, we examined the effects of the chlorophyll metabolite pyropheophorbide-a (P-a) on ATP synthesis in isolated mouse liver mitochondria in the presence of red light (λmax = 670 nm), which chlorin-type molecules such as P-a strongly absorb (Aronoff, 1950), and to which biological tissues are relatively transparent. We used P-a because it is an early metabolite of chlorophyll, however, most known metabolites of chlorophyll can be synthesized from P-a by reactions that normally take place in animal cells. Control samples of mitochondria without P-a, and/or kept in the dark were also assayed. In the presence of P-a, mitochondria exposed to red light produce more ATP than mitochondria without P-a (Fig. 1A) or mitochondria kept in the dark (supplementary material Fig. S1A–D). Mitochondrial membrane potential (Fig. 1B) and oxygen consumption (Fig. 1C) increased upon increased light exposure in P-a-treated mitochondria. Light or P-a alone had no effect on any of the above measures of mitochondrial activity (supplementary material Fig. S1E–G). With too much added P-a, ATP concentrations and the rate of oxygen consumption started to return to the levels in mitochondria not incubated with P-a (supplementary material Fig. S1G). Addition of the electron transport inhibitor, sodium azide, reduced the light- and P-a-fueled oxygen consumption by 57% (supplementary material Fig. S1H–I), consistent with oxygen consumption occurring through the electron transport system. Observations were consistent with enhanced ATP production driven by oxidative phosphorylation.
Fig. 1.Chlorophyll metabolite P-a allows isolated mouse liver mitochondria to capture light to make ATP. (A) ATP synthesis in mouse liver mitochondria incubated with P-a (treated) and exposed to light compared to controls (no P-a). Light exposure started at time zero and ADP was added at 30 seconds. Aliquots were obtained at times shown and relative ATP levels measured using the firefly luciferase assay. Means and standard deviations are shown for each time point. The experiment was run in triplicate with the same batch of mitochondria. *P<0.05 for treated versus control samples. (B) Mitochondrial membrane potential (Δψm) under different treatments as measured by safranin fluorescence. Lower fluorescence equals higher membrane potential. Mitochondria, with or without P-a, were exposed to light for 2 minutes or kept in the dark. Safranin was added at time zero and safranin fluorescence was continuously measured while samples remained under the light. The experiment was run in triplicate with the same batch of mitochondria. Curves shown are the average traces for triplicate runs. (C) Representative oxygraph trace (black line) for mitochondria treated with 4 µM P-a. The light was turned on or off at the times indicated by the arrows. Steeper slope denotes faster oxygen consumption. Dotted lines show slopes when the light was off. When the light was turned on the slope of the black line increased by twofold. That is, oxygen consumption increased when the light was turned on. When the light was turned off, oxygen consumption returned to baseline levels (i.e. the two gray lines have the same slope).
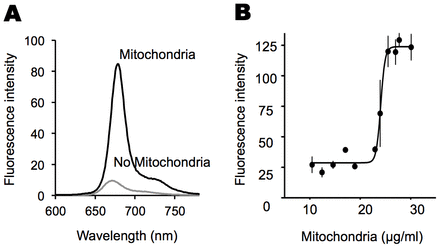
To determine whether P-a associates with mitochondria, we measured P-a fluorescence at 675 nm in the presence of increasing amounts of heart mitochondrial fragments obtained from sheep (Fig. 2A,B). After increasing the concentration of mitochondria, P-a fluorescence increased abruptly, by fivefold, and quickly reached a plateau (Fig. 2B). The abrupt change in fluorescence reflects a change in the environment of P-a, consistent with its change from an aqueous environment to one in which it is presumably associated with a protein. This threshold-sensitive behavior is consistent with zero-order ultrasensitivity, or positively cooperative binding, as described by Goldbeter and Koshland, and suggests a coordinated interaction between the metabolite and mitochondrial fragments (Goldbeter and Koshland, 1981). In contrast, this threshold sensitivity was not observed when increasing amounts of bovine serum albumin (BSA) were added to a solution of P-a; instead, fluorescence steadily increased (supplementary material Fig. S1J).
Fig. 2.Cooperative binding of P-a to mitochondrial fragments. (A) Fluorescence spectra of P-a before and after addition of sheep heart mitochondrial fragments. Upon addition of mitochondrial fragments, the fluorescence intensity of P-a increased and shifted to a longer wavelength, and the shape of the curve (ratio of the shoulder to main peak) changed. (B) Ultrasensitive steady state response of the P-a–mitochondrial interaction. We measured fluorescence intensity for a 1 µM P-a solution while increasing the concentration of mitochondrial fragments. A Hill coefficient of 36, with a 95% confidence interval from 7 to 65, was obtained by fitting the data to the Hill equation [y = axb/(cb+xb)+offset]. Fit (R2): 0.96.
Catabolic reduction of coenzyme Q10 (CoQ10) is a rate limiting step in respiration (Crane, 2001). The majority of CoQ10 molecules exist in two alternate states of oxidation: ubiquinone, the oxidized form, and ubiquinol, the reduced form. To show that the P-a metabolite could catalyze the photoreduction of mitochondrial CoQ10, we measured the oxidation state of CoQ10 in the above sheep heart mitochondrial fragments in response to exposure to red light. We exposed the mitochondria to light for 10 minutes and measured the percentage of reduced and oxidized CoQ10 by high performance liquid chromatography (HPLC) (Qu et al., 2013). In the freshly isolated mitochondria fragments, nearly all the CoQ10 was oxidized in the form of ubiquinone. However, when we incubated the mitochondria with P-a and exposed the suspension to light, 46% of CoQ10 was reduced (Table 1, entry 1). In comparison, as a positive control, we energized the mitochondria with glutamate/malate and kept the suspension in the dark, yielding a 75% reduction of CoQ10 within 10 minutes (entry 2). In the absence of light, no reduction occurred (entry 3). Upon denaturing the mitochondrial proteins with heat, no reduction occurred (entry 4). Likewise, there was a lack of CoQ10 reduction with CoQ10, P-a and light in the absence of mitochondria (entry 5). These observations are consistent with the fluorescence data in Fig. 2A,B, showing that mitochondrial proteins sequester and organize P-a. In the absence of added P-a, a 2–14% reduction was observed, depending on the mitochondrial preparation used (entry 6). We attribute this ‘background reduction’ to the actions of endogenous chlorophyll metabolites, which we were able to detect by fluorescence spectroscopy (see Distribution of light-absorbing dietary chlorophyll, below).
Table 1. Photoreduction of CoQ10 is an early event in light-stimulated ATP synthesisLight-driven ATP synthesis in rodent tissue homogenates
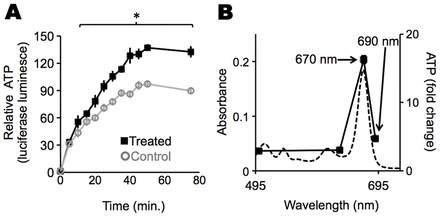
To determine whether chlorophyll metabolites and light could influence ATP production in whole tissues, we treated mouse brain homogenates with P-a and exposed them to 670-nm light. The treated brain homogenates synthesized ATP at a 35% faster rate than a control homogenate that was not incubated with P-a [relative ATP synthesis rates (means with standard error and 95% confidence intervals (CI) were: treated, 171.7±8.1 (CI: 154.6–188.7); control, 111.3±9.1 (CI: 92.5–130.0); Fig. 3A]. No linear correlation between the increase in ATP concentrations and the amount of added P-a was observed. Increasing concentrations of P-a elicited the same increase in ATP (supplementary material Fig. S2A,B).
Fig. 3.Chlorophyll metabolite P-a allows mouse brain tissue homogenates to capture light to make ATP. (A) ATP synthesis in mouse brain homogenate with light exposure. Homogenates were incubated with ADP ± P-a and exposed to light starting at time zero. Aliquots were withdrawn at the times shown. Relative ATP in the aliquots was measured using the firefly luciferase assay. The experiment was run in triplicate with the same batch of homogenized brains. Means and standard errors are shown for each time point. For the control, the standard errors are smaller than the line markings and thus cannot be seen. *P<0.05 for treated versus untreated samples. (B) Overlay of the absorption spectrum of P-a (dotted line) and the wavelengths tested for ATP production in samples treated with P-a and exposed to light for 20 minutes. Peak ATP production correlated with peak P-a absorption. Experiments were done in triplicate. Means and standard errors were calculated, however, standard errors are smaller than the markings and thus cannot be seen.
To demonstrate that photon absorption by P-a was necessary to enhance ATP production, we exposed the P-a-treated brain homogenates to greenish (500 nm) and red (630, 670 and 690 nm) light, all with the same total energy. Wavelengths of light that were more strongly absorbed by P-a produced the largest increase in ATP. For example, the ATP concentration increased by ∼16-fold during exposure to 670 nm light; relative to the same sample kept in the dark, it increased by two-to-fivefold during exposure to 500, 630 and 690-nm light of equal energy (Fig. 3B).
In addition to brain homogenates, P-a also enhanced ATP production in adipose, lens and heart homogenates (supplementary material Fig. S2C–E). Quantification of ATP by both the luciferase assay and high-performance liquid chromatography (HPLC) gave similar results (supplementary material Fig. S2E–F).
Distribution of light-absorbing dietary chlorophyll
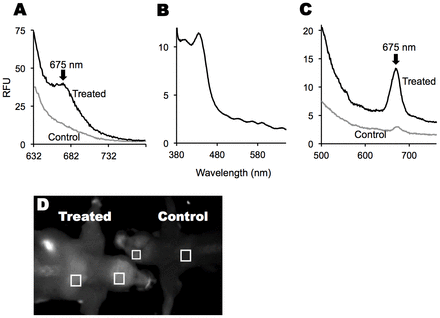
Chlorophylls and its metabolites, both chlorins, have signature absorption and admission spectra (Aronoff, 1950). Namely they absorb strongly (ε≈50,000 M−1 cm−1) at ∼665–670 nm and demonstrate intense fluorescence emissions at ∼675 nm, which differentiate chlorins from endogenous molecules in mammals (Aronoff, 1950). To examine whether dietary chlorophyll and/or its metabolites were present in animal tissue after oral consumption, we fed mice a chlorophyll-rich diet. Brain (Fig. 4A) and fat (Fig. 4C) extracts from these mice exhibited red fluorescence at 675 nm when excited with a 410-nm light [brain: treated, 15.4±6.7 (n = 6); control: 4.2±2.6 (n = 6; means ± s.d.); P<0.01]. The excitation spectrum of this 675-nm peak (Fig. 4B) was similar to that of known chlorophyll metabolites with an intact chlorin ring: with maxima at 408, 504, 535, 562 and 607 nm. This red fluorescence diminished, as measured by the area under the 675 nm peak, when animals were given a chlorophyll-free diet for 2 weeks. Red fluorescence could also be seen using fluorescence imaging; fluorescence was stronger in the bodies and brains of animals fed chlorophyll than in animals given a chlorophyll-poor diet [Fig. 4D; mean gray value in the boxed areas with standard deviation and minimum and maximum gray value shown in brackets were: treated brain, 118 (97–138); control brain, 82 (60–100); treated back fat pad, 116 (97–132) and control back fat pad, 35 (25–46)]. The red fluorescence was enriched in the gut and intestines, consistent with dietary chlorophyll being the source of the fluorescence.
Fig. 4.Dietary chlorophyll results in chlorophyll-metabolite-like fluorescence in tissues. (A) Representative fluorescence spectra of brain extracts following excitation at 410 nm. Relative peak areas for a total of six control animals fed a chlorophyll-poor diet and six treated animals fed a chlorophyll-rich diet. (B) Representative excitation spectrum (emission at 675 nm) of a brain extract from mice fed a chlorophyll-rich diet. (C) Representative fluorescence spectra of abdominal fat extracts from mice fed chlorophyll-poor and rich diets. (D) A 675±10-nm fluorescence image of skinned mice raised on chlorophyll-rich and -poor diets.
To determine whether the red fluorescence was localized to mitochondria, we measured the relative 675-nm fluorescence in whole liver homogenates and mitochondria isolated from these homogenates. As measured by fluorescence intensity, isolated mitochondria contained 2.3-fold as much of the 675-nm fluorescent metabolite(s) per milligram of protein as did the whole liver homogenate. This observation suggests that P-a was concentrated in the mitochondria, consistent with data summarized in Fig. 2A,B, and literature reports (MacDonald et al., 1999; Tang et al., 2006).
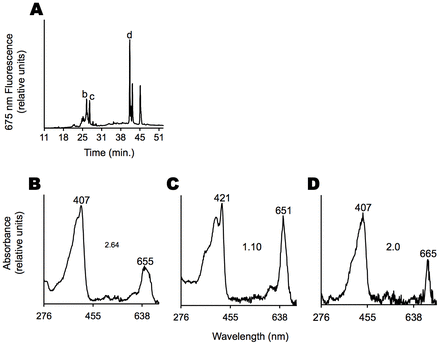
Fat and plasma extracts from rats fed chlorophyll-rich diets were further analyzed by HPLC to elucidate the source of the red 675-nm fluorescence. Fig. 5A shows a representative chromatogram with compounds in the eluting solvent that displayed 675-nm fluorescence when excited with 410-nm light. Rat fat extracts and plasma extracts both contained similar chlorophyll-derived metabolites (similar chromatograms not illustrated). Two groups of compounds eluting at 23–30 minutes and 40–46 minutes were detected. Compounds eluting between 23 and 30 minutes had similar retention times to those of the chlorophyll metabolites without the phytyl tail, with at least one carboxylate group, such as P-a. The absorption spectra (the locations of the absorbance maxima and the Soret-to-Qy-band ratios) of this group of compounds were consistent with demetalated chlorophylls (Rabinowitch, 1944), as shown in Fig. 5B. In addition, the spectra of this group of peaks were indicative of coordination to a metal ion. A representative spectrum of such a presumably metalated metabolite is shown in Fig. 5C, showing a red shifted Soret band, a blue shifted Qy-band and a Soret-to-Qy-band ratio of ∼1. The compounds eluting between 40 and 46 minutes had similar retention times to that of the demetalated chlorophyll-a standard (pheophytin-a). In addition, these compounds partitioned with hexanes (polarity index = 0.1) when mixed with hexanes and acetonitrile (polarity index = 5.8). This latter characteristic is consistent with a lack of a carboxylic acid group, or an esterified P-a, such as pheophytin-a. Similar HPLC chromatograms from fat extracts of swine fed chlorophyll rich diets (Mihai et al., 2013) were recorded (supplementary material Fig. S2G), suggesting that uptake and distribution of chlorophyll metabolites were not unique to mice and rats.
Fig. 5.Light-absorbing metabolites of chlorophyll are present in adipose tissue. (A) HPLC chromatogram of an adipose extract. 2.5 grams of abdominal adipose tissue from a rat fed a chlorophyll-rich diet was extracted with acetone and the acetone concentrate subjected to HPLC. In the chromatogram, only compounds that displayed 675-nm fluorescence, characteristic of chlorophyll and its metabolites possessing a chlorin ring, are shown. Five major peaks are observed along with several minor peaks. For peaks with letters, the corresponding absorption spectra are shown below. (B–D) Absorption spectra of labeled peaks in A (b–d, respectively). Numbers above peaks are peak maxima in nm. Numbers in the center are the ratios of the Soret band, around 400 nm, to the Qy band at around 655 nm. All spectra are consistent with those of metabolites of chlorophyll. Spectrum C has been assigned to a metalated porphyrin.
We quantified total blood pigments from rats that absorbed at 665 nm. Using an extinction coefficient of 52,000 at 665 nm (Lichtenthaler, 1987), which is typical of chlorophyll-a-derived pheophytins, we estimated a plasma concentration of 0.05 µM in two rats fed a chlorophyll-rich diet. The 665-nm peak was absent in animals fed a chlorophyll-poor diet. The amount of measured total metabolite was five- and two-times higher than that reported for the fat soluble vitamins K (Tovar et al., 2006) and D (Halloran and DeLuca, 1979), respectively, in the rat.
Light-driven ATP synthesis in C. elegans
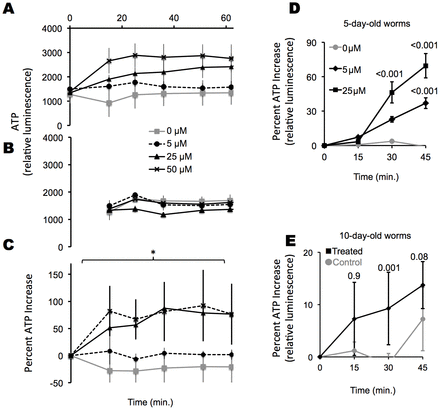
Next, we used C. elegans to evaluate the effects of light-stimulated ATP production in a complex organism. As C. elegans age, there is a drop in cellular ATP (Braeckman et al., 1999; Braeckman et al., 2002). We hypothesized that the worm would live longer if it could offset this decline in ATP by harvesting light energy for ATP synthesis. As our model system, we used firefly luciferase-expressing C. elegans, which upon incubation with luciferin emit a luminescence that is proportional to their ATP pools (Lagido et al., 2009; Lagido et al., 2008; Lagido et al., 2001). Upon incubation with P-a, worms incorporated the metabolite, as measured by fluorescence spectroscopy (supplementary material Fig. S3A). To determine whether there were changes in ATP stores in response to light, we plated two groups of worms into 96-well plates containing luciferin substrate. We measured worm luminescence at time zero. We then exposed one group to 660-nm light and kept the other in the dark and periodically measured luminescence in both groups of worms (summarized in Fig. 6A,B). To determine whether ATP increased in light-exposed animals, we subtracted the luminescence signal of the worms kept in the dark from that of the worms exposed to light (Fig. 6C). Worms that were given P-a had a statistically significant increase in ATP when exposed to light, whereas control worms showed no increase. The metabolite alone had no effect on ATP levels when the worms were kept in the dark (i.e. luminescence intensity remained constant throughout the experiment). The elevated luminescence signal persisted for 1 hour after the light was turned off, at which time measurement ceased. However, the luminescence intensity did not further increase during the time the light was off. It was unclear whether this persistent signal reflected the kinetics of the luciferase–luciferin reaction, luciferase expression, or actual ATP pools. Thus ATP was quantified by additional methods.
Fig. 6.P-a treatment enables worms to capture light to generate ATP. Black lines show results from worms incubated with P-a at the indicated concentrations; gray lines show results from worms not incubated with P-a. (A) In vivo, real-time ATP levels in 1-day-old worms were tracked during exposure to light. Luciferase-expressing worms were incubated with luciferin and exposed to light at time zero. Luminescence was measured at the times shown. Data represent triplicate experiments of 12 separate sets of worms plated in 12 wells of a 96-well pate. Means and standard deviations are shown for each of the three separate runs. (B) In vivo, real-time ATP levels in worms kept in the dark. The same experiment as in A in the same 96-well plate, but the worms were kept in the dark. (C) Percentage ATP increase for worms in A relative to worms in B. (D) In vivo, real-time ATP monitoring. Groups of worms were incubated with or without P-a; light exposure began at time zero and in vivo ATP levels were determined at the times shown in each group of worms by measuring worm luminescence after the addition of luciferin. Each time point represents a different group of worms exposed to light for the times shown. Each experiment was performed in triplicate sets of 12; averages and standard deviations are shown. P-values of Student’s t-tests are also shown, representing the significance compared with the controls at the same light exposure. (E) The same experiment as described in D, but using 10-day-old worms.
As an alternative means of determining whether light stimulated ATP synthesis, we plated luciferase-expressing worms into a 96-well plate without the luciferin substrate, and exposed them to light. ATP status was determined at time zero, immediately before light exposure, and at 15-minute intervals for a total of 45 minutes by adding the luciferin substrate to a group of worms and measuring luminescence (Fig. 6D,E). We found an increase in ATP when 5-day-old and 10-day-old adult worms were fed the metabolite and exposed to light.
We further confirmed the in vivo increase in ATP using two additional ex vivo methods. After light treatment, we lysed the worms, extracted their ATP and quantified ATP in the homogenate using either the firefly luciferase assay or HPLC (supplementary material Fig. S3B,C). Both methods were consistent with the in vivo ATP measurements.
In addition to an increase in ATP, worms treated with P-a exhibited a 13% increase in respiration when exposed to light, as measured by oxygen consumption. However, light had no effect on the respiration rates in untreated worms (supplementary material Fig. S3D). This observation is consistent with an increase in ATP through oxidative phosphorylation, in accordance with the mitochondrial data. Despite the increase in ATP, the levels of reactive oxygen species (ROS) were equivalent in treated and untreated worms during 5 hous of light exposure, as measured using 2′,7′-dichlorofluorescin diacetate (supplementary material Fig. S3E). In fact, although the difference was not statistically significant, treated worms exhibited, on average, lower levels of ROS.
Light harvesting to extend life span
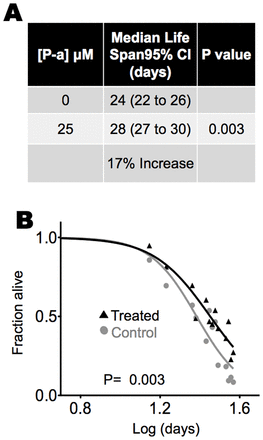
We next tested whether photonic energy absorption by P-a could prolong life. Life span measurements were taken in liquid cultures according to the method of Gandhi et al. and Mitchell et al. (Gandhi et al., 1980; Mitchell et al., 1979). Adult worms were incubated with P-a for 24 hours. Beginning at day 5 of adulthood, we exposed the worms to red light in a daily 5 hours:19 hours light∶dark cycle. Control worms were not given P-a or exposed to light, but otherwise were kept under identical conditions. Counts were made at 2- to 3-day intervals and deaths were assumed to have occurred at the midpoint of the interval. To obtain the half-life, we plotted the fraction alive at each count verses time and fitted the data to a two-parameter logistic function, known to accurately fit survival of 95% of the population (Vanfleteren et al., 1998). The group treated with P-a and light had a 17% longer median life span than the group that was not treated with P-a, but exposed to light (Fig. 7A,B). P-a treatment alone, in the absence of light, had no effect on life span (supplementary material Fig. S4B). Light treatment alone decreased life span by 10% (supplementary material Fig. S4B), in accordance with reports that nematodes survive better in complete darkness (Thomas, 1965). This decrease in median life span brought on by light was reversed when the worms were treated with P-a. The increased median life span with light and P-a was reproducible with different batches of worms (supplementary material Fig. S4B–E). Increasing the amount of P-a past a certain threshold, however, lead to a gradual decrease in lifespan approaching that of animals not treated with P-a (supplementary material Fig. S4B,C).
Fig. 7.P-a and light increase C. elegans median life span. (A) Median life spans of worms treated with P-a and exposed to light versus those exposed to light but not treated with P-a. Numbers in parentheses are 95% confidence intervals (CI). (B) Life span plots of the values used for A. P-value is from an f-test. Experiments were run in triplicate. The L4 molt was used as time zero for life span analysis. Worms were grown in liquid culture at 500 worms/ml. For counting, aliquots were withdrawn and placed in a 96-well plate to give ∼10 worms per well; the worms were scored dead or alive on the basis of their movement, determined with the aid of a light microscope. A total of 60–100 worms, representing 1–2% of the total population, were withdrawn and counted at each time point for each flask.
We also examined life span longitudinally. We placed 6-day-old adult P-a- and non-P-a-treated worms into a 96-well plate, exposed them to red light for 5 hours per day and compared the percentage dead and alive after 15 days. Result: 47% of the P-a-treated worms were alive (175 alive; 200 dead) after 15 days, versus 41% of the control worms (111 alive; 163 dead), consistent with the cross-sectional experiments above.
DISCUSSION
Photoreduction of coenzyme Q
Upon incubation of: (1) isolated mouse mitochondria; (2) mouse brain, heart and lens homogenates; (3) homogenized duck fat; and (4) live C. elegans, with a representative metabolite of chlorophyll, light exposure was able to increased ATP concentrations. These observations in a variety of animal tissues perhaps demonstrate the generality of this phenomenon. To synthesize ATP, mitochondrial NADH reductase (complex I) and succinate reductase (complex II) extract electrons from NADH and succinate, respectively. These electrons are used to reduce mitochondrial CoQ10, resulting in ubiquinol (the reduced form of CoQ10). Ubiquinol shuttles the electrons to cytochrome c reductase (complex III), which uses the electrons to reduce cytochrome c, which shuttles the electrons to cytochrome c oxidase (complex IV), which ultimately donates the electrons to molecular oxygen. As a result of this electron flow, protons are pumped from the mitochondrial matrix into the inner membrane space, generating a trans-membrane potential used to drive the enzyme ATP-synthase.
The ‘pool equation’ of Kröger and Klingenberg describes the total rate of electron transfer: Vobs = VoxVred/(Vox+Vred), where Vred is ubiquinone reduction and Vox is ubiquinol oxidation (Kröger and Klingenberg, 1973). Based on this equation, the major roles of complexes I and II can be considered to maintain the mitochondrial ubiquinol pool, and to reduce ubiquinone, which should result in increased ATP synthesis. We reasoned the reduction of CoQ10 could be a potential step in the respiratory pathway in which chlorophyll metabolites could influence ATP levels, as it is known that chlorophyll-type molecules can photoreduce quinones (Chesnokov et al., 2002; Okayama et al., 1967). Indeed, a primary step during photosynthesis is the reduction of the quinone, plastoquinone, by a photochemically excited chlorophyll a (Witt et al., 1963). We hypothesized that if the reduction of mitochondrial ubiquinone could be catalyzed by a photoactivated chlorophyll metabolite, such as P-a, then ATP synthesis would be driven by light in mitochondria with these dietary metabolites. In the proposed mechanism, electrons would be transferred by a metabolite of chlorophyll to CoQ10, from a chemical oxidant present in the mitochondrial milieu. Many molecules, such as dienols, sulfhydryl compounds, ferrous compounds, NADH, NADPH and ascorbic acid, could all potentially act as electron donors. Throughout mammalian evolution, photons of red light from sunlight have been present deep inside almost every tissue in the body. Photosensitized electron transfer from excited chlorophyll-type molecules is widely hypothesized to be a primitive form of light-to-energy conversion that evolved into photosynthesis (Krasnovsky, 1976). Thus it is tempting to speculate that mammals possess conserved mechanisms to harness photonic energy.
Photoexcitation of chlorophyll and derivatives produces the excited singlet state (*1). Oxidative quenching of this singlet state by ubiquinone is possible. Electron transfer could take place through proteins or in solution. Escape from the charge transfer complex and protonation would yield ubisemiquinone, which accounts for 2–3% of the total ubiquinone content of mitochondria (De Jong and Albracht, 1994). Ubisemiquinone can be reduced to ubiquinol by repeating the above sequence or by disproportionation to give one molecule of ubiquinol and one molecule of ubiquinone. Back-electron transfer, from the photoreduced metabolite to the oxidized quinone, could be inhibited by disproportionation or by organizing the chlorophyll derivative and ubiquinol through protein binding. In line with the CoQ10 photoreduction hypothesis, we observed mitochondrial CoQ10 was reduced when isolated mitochondria were exposed to light and P-a (Table 1). Also consistent with light and/or P-a acting upstream of complexes I and II, in isolated mitochondria we observed an increase in ATP in the absence of added electron transport substrates, such as glutamate and malate (Fig. 1A; supplementary material Fig. S1A–C). However, further evidence is needed to confirm this mechanistic hypothesis.
The effect of light in vivo
Intense red light between 600 and 700 nm has been reported to modulate biological processes (Hashmi et al., 2010; Passarella et al., 1984; Wong-Riley et al., 2005), and has been investigated as a clinical intervention to treat a variety of conditions (Hashmi et al., 2010). Exposure to red light is thought to stimulate cellular energy metabolism and/or energy production by, as yet, poorly defined mechanisms (Hashmi et al., 2010). In the presence of P-a, we observed changes in energetics in animal-derived tissues initiated with light of intensity and wavelengths (≈670 nm at ≈0.8±0.2 W/m2) that can be found in vivo when outdoors on a clear day. On a clear day the amount of light illuminating your brain would allow you to comfortably read a printed book (Benaron et al., 1997). In humans, the temporal bone of the skull and the scalp attenuate only 50% of light at a wavelength of ∼670 nm (Eichler et al., 1977; Wan et al., 1981). In small animals, light can readily reach the entire brain under normal illumination (Berry and Harman, 1956; Massopust and Daigle, 1961; Menaker et al., 1970; Vanbrunt et al., 1964). Sun or room light over the range of 600–700 nm can penetrate an approximately 4-cm-thick abdominal wall with only three-to-five orders of magnitude attenuation (Bearden et al., 2001; Wan et al., 1981). Photons between 630 and 800 nm can penetrate 25 cm through tissue and muscle of the calf (Chance et al., 1988). Adipose tissue is bathed in wavelengths of light that would excite chlorophyll metabolites (Bachem and Reed, 1931; Barun et al., 2007; Zourabian et al., 2000). Thus, identification of pathways, which might have developed to take advantage of this photonic energy, may have far-reaching implications.
Dietary chlorophyll in animals
A potential pathway for photonic energy capture is absorption by dietary-derived plant pigments. Little is known about the pharmacokinetics and pharmacodynamics of dietary chlorophyll or its chlorin-type metabolites in human tissues. Here, we observed the accumulation of chlorin-type molecules in mice, rats and swine administered a diet rich in plant chlorophylls (Figs 4, 5; supplementary material Fig. S2G). Data suggests that sequestration from the diets of chlorophyll-derived molecules, which are capable of absorbing ambient photonic energy, might be a general phenomenon.
To date, the reported chlorophyll metabolites isolated from animals have been demetalated (Egner et al., 2000; Fernandes et al., 2007; Scheie and Flaoyen, 2003). The acidic environment of the stomach is thought to bring about loss of magnesium from the chlorophyll (Ferruzzi and Blakeslee, 2007; Ma and Dolphin, 1999). Our absorbance data from extracted pigments from rat fat is consistent with the presence of chlorophyll metabolites bonded to a metal (Fig. 5). If true, the presence of a metal derivative in fat tissue suggests that the pigment was actively re-metalated to take part in coordination chemistry. The identification of several metabolites in the fat and plasma of rats and swine fed a chlorophyll-rich diet that are similar to ones found in plants is significant. However, the structures of the metabolites remain to be elucidated. Chlorin-type molecules are similar in structure and photophysical properties and thus can carry out similar photochemistry (Gradyushko et al., 1970). Our data demonstrate that dietary metabolites of chlorophyll can be distributed throughout the body where photon absorption may lead to an increase in ATP as demonstrated for the chlorin P-a. Indeed, P-a could have been transformed into other metabolites, as most known metabolites of chlorophyll can be formed from P-a by reactions that normally take place in animal cells.
There relationship between the increase in ATP and the amount of added P-a was not linear (supplementary material Fig. S2A,B). ATP stimulation by light in the presence of P-a better fitted a binary on/off, rather than a graded response to P-a. Increasing concentrations of P-a elicited the same increase in ATP, after light exposure. However, with too much added P-a, ATP levels began to fall. This on/off response was also consistent with the observed cooperative binding mode of P-a with mitochondria fragments, suggesting that the threshold response may be regulated by mitochondrial binding of P-a. If chlorophyll metabolites are found to be involved in energy homeostasis, a better understanding of their pharmacodynamics and pharmacokinetics will be needed.
ATP stores and life span
Light of 670 nm wavelength that penetrates the human body, yields ∼43 kcal/mol (1.18×10−22 kcal/photon). Given estimated concentrations of chlorophyll derivatives in the body (Egner et al., 2000; Fernandes et al., 2007; Scheie and Flaoyen, 2003) and the photon flux at 670 nm (Bachem and Reed, 1931; Barun et al., 2007; Bearden et al., 2001; Benaron et al., 1997; Chance et al., 1988; Eichler et al., 1977; Menaker et al., 1970; Vanbrunt et al., 1964; Wan et al., 1981; Zourabian et al., 2000), each chlorophyll metabolite would be expected to absorb only a few photons per second. As such, one might anticipate negligible amounts of additional energy. Organization of chlorophyll metabolites into supramolecular structures, similar to chlorophyll antenna systems in photosynthetic organisms, would increase the effective cross-sectional area of photon absorption and, thus, photon catch. Indeed, our observed positively cooperative binding with mitochondrial fragments is evidence for such organization. Even so, to approach the rate of ATP synthesis powered by NADH or FADH2, sufficient P-a pigment would have to be added to turn animals green. Nevertheless, in model systems, we measure an increase in ATP upon light absorption and changes in fundamental biology (extention in life span). Regardless of the mechanism by which ATP is increased or the measured amount of the increase, perhaps the larger question is: how much of an increase in ATP is enough to make a biological difference?
In animals, treatment with P-a and light both increased ATP and median life span, suggesting that light in the presence of these light absorbing dietary metabolites can significantly affect fundamental biological processes. We previously observed that chlorophyll metabolites enabled photonic energy capture to enhance vision using a mouse model (Isayama et al., 2006; Washington et al., 2004; Washington et al., 2007). Because ATP can regulate a broad range of biological processes, we suspect that ATP modulation also played a role in vision enhancement. The increase in life span may seem contradictory, given that there are studies suggesting that limiting metabolism and ATP synthesis increases the life span of C. elegans. It has been proposed that the life span of this worm might be determined by the metabolic status during development (Dillin et al., 2002) and that there might be a coupling of a slow early metabolism and longevity (Lee et al., 2003). Other observations have led to the hypothesis that increased life span may be achieved by decreasing total energy expenditure across the worm’s entire life span (Van Raamsdonk et al., 2010). However, most studies decrease ATP synthesis from hatching through genetic engineering. By contrast, here, we were able to increase ATP during adulthood at a time when ATP stores reportedly begin to decline. For example, by day 4 of adulthood, the level of ATP and oxygen consumption can drop by as much as 50% compared to day zero (Braeckman et al., 1999; Braeckman et al., 2002). This difference in timing might account for why we observed an increase in life span in response to an increase in ATP. We note that besides caloric restriction, there are only a few interventions that are known (Petrascheck et al., 2007) to increase life span when given to an adult animal.
Alternative mechanisms of life-span extension cannot be ruled out. For example, an increase in reactive oxygen species (ROS) is thought to increase life span in C. elegans (Heidler et al., 2010; Schulz et al., 2007). Upon photon absorption, metabolites of chlorophyll can transfer energy to oxygen, resulting in the generation of singlet oxygen, a ROS. Thus life-span extension seen here might be a result of an increase in ROS due to the generation of singlet oxygen. However, our published data with blood plasma (Qu et al., 2013) and data here from C. elegans do not show an increase in ROS. As ubiquinol is a potent lipid antioxidant (Frei et al., 1990) any ROS increase might be offset by an increase in ubiquinol, generated from the photoreduction of coenzyme Q. Indeed, by producing ubiquinol, P-a might have increased life span by an alternative method by protecting against long-term oxidative damage, which is also a mechanism that has been shown to increase C. elegans life span (Ishii et al., 2004). Further research will be needed to distinguish between the above possible mechanisms.
Conclusion
Both increased sun exposure (Dhar and Lambert, 2013; John et al., 2004; Kent et al., 2013a; Kent et al., 2013b; Levandovski et al., 2013) and the consumption of green vegetables (Block et al., 1992; Ferruzzi and Blakeslee, 2007; van’t Veer et al., 2000) are correlated with better overall health outcomes in a variety of diseases of aging. These benefits are commonly attributed to an increase in vitamin D from sunlight exposure and consumption of antioxidants from green vegetables. Our work suggests these explanations might be incomplete. Sunlight is the most abundant energy source on this planet. Throughout mammalian evolution, the internal organs of most animals, including humans, have been bathed in photonic energy from the sun. Do animals have metabolic pathways that enable them to take greater advantage of this abundant energy source? The demonstration that: (1) light-sensitive chlorophyll-type molecules are sequestered into animal tissues; (2) in the presence of the chlorophyll metabolite P-a, there is an increase in ATP in isolated animal mitochondria, tissue homogenates and in C. elegans, upon exposure to light of wavelengths absorbed by P-a; and (3) in the presence of P-a, light alters fundamental biology resulting in up to a 17% extension of life span in C. elegans suggests that, similarly to plants and photosynthetic organisms, animals also possess metabolic pathways to derive energy directly from sunlight. Additional studies should confirm these conclusions.
MATERIALS AND METHODS
General procedures
Two light sources were used for all experiments, either a 300 W halogen lamp equipped with a variable transformer and band pass interference filters [500, 632, 670, 690 nm with full-width half maximum (FWHM) of 10 nm] or a 1.70 W, 660 nm, LED light bulb. Luminous power density was set to 0.8±0.2 W/m2 as measured by a LI-250A light meter (LI-COR Biosciences, Lincoln, NE). The intensity of red light used was 30–60 times less than the level of red light that we measured on a clear March afternoon in New York City and is less than the level that several organs are exposed to in vivo. Pyropheophorbide-a (P-a, 95% purity) was obtained from Frontier Scientific, Logan, UT. For all experiments, prior to exposing samples to light, we minimized light exposure by preparing samples/experiments with laboratory lights turned off, using a minimum amount of indirect sunlight that shone through laboratory windows (>0.001 W/m2).
Animal protocols were approved by the Institutional Animal Care and Use Committee of Columbia University. Mice (ICR, Charles River, Wilmington, MA) weighing 22–28 g and rats (Fisher 344, Harlan Teklad, Indianapolis, IN), weighing 300 g were used. Swine, fed a chlorophyll-rich diet have been described previously (Mihai et al., 2013).
Continuous ATP monitoring in isolated mouse liver mitochondria
Mice were fed a chlorophyll-poor, purified rodent diet supplied by Harlan (Indianapolis, IN) for a minimum of 2 weeks. We isolated mouse liver mitochondria by differential centrifugation according to existing procedures (Frezza et al., 2007) and used only preparations with a minimum respiratory control ratio above 4.0 [state III/II, using glutamate (5 mM final) and malate (2.5 mM final) as measured with an oxygen electrode from Qubit Systems Inc., Kingston, ON, Canada]. Mitochondria at a final concentration of ≈1 mg protein/ml as determined by the Coomassie Plus (Bradford) protein assay (Thermo Fisher Scientific, Rockford, IL) in buffer A (0.250 M mannitol, 0.02 M HEPES, 0.01 M KCl, 0.003 M KH2PO4, 0.0015 M MgAc2·H2O, 0.001 M EGTA, 1 mg/ml fatty acid–poor BSA, pH 7.4) were incubated with P-a for 30 minutes at 0°C. ADP was added (0.5 mM final concentration) and then 250 µl aliquots of this suspension were placed in nine wells of a 96-well plate for exposure to light at room temperature. At various times, 20 µl aliquots were withdrawn, added to 150 µl lysis buffer (10 mM Tris, pH 7.5; 100 mM NaCl; 1 mM EDTA and 1% Triton X-100), and ATP levels were determined with a commercial kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Controls were treated in the same way, except they were: (1) incubated at 0°C without P-a (shown), (2) not exposed to light and (3) were incubated without P-a and not exposed to light.
Membrane potential measurement
Mitochondrial membrane potential was monitored in buffer A as described by Feldkamp et al. (Feldkamp et al., 2005). Measurements were made in a 3 ml cuvette placed inside a fluorescence spectrometer (Fluorormax-4, HORIBA Jobin Yvon, Horiba Scientific, Kyoto, Japan) with a final reaction volume of 1 ml. For light exposure, we used a fiber optic light guide to capture and direct light from a 660 nm LED light bulb into the spectrometer. The end of the fiber optic cable was positioned 1 cm above the reaction mixture. Prior to these experiments, light power was measured 1 cm from the end of the fiber optic cable.
Oxygen consumption measurement
Mitochondrial oxygen consumption was measured using an oxygen electrode cuvette (OX1LP-1 ml; Qubit Systems Inc., Kingston, ON, Canada) according to the manufacturer’s instructions. Reactions were run with mitochondria at a concentration of ≈1 mg protein/m’ in buffer A. For light exposure, a 660-nm LED was directed at the plastic [poly-(methyl methacrylate)] chamber.
For inhibition of respiration, sodium azide was added at a final concentration of 0.005 M from a stock solution in water. Sodium azide inhibits cytochrome oxidase (complex IV): oxygen consumption during state 3 respiration is progressively inhibited by increasing concentrations of azide (Bogucka and Wojtczak, 1966).
Analysis of zero-order ultrasensitivity
Mitochondria from sheep hearts were prepared as previously described (Smith, 1967) on two separate occasions from 2 and 1 sheep heart(s) using ‘Procedure 1’. We used mitochondrial fragments to allow P-a direct access to the respiratory chain, to minimize potential complications due to variable rates of P-a import. Mitochondrial isolation started within 1 hour of the death of the animal and the hearts were transported to the laboratory in a bath of 0.25 M sucrose, 0.1 M tris(hydroxymethyl)aminomethane (Tris) at pH 7.5, which was surrounded by ice. Mitochondria were isolated and stored in 250 µl aliquots at a concentration of ∼60 mg of protein/ml in 300 mM trehalose, 10 mM HEPES–KOH pH 7.7, 10 mM KCl, 1 mM EGTA, 1 mM EDTA and 0.1% BSA at −80°C (Yamaguchi et al., 2007) until use. The thawed mitochondria exhibited a respiratory control ratio of ∼1, indicating mitochondrial fragmentation.
Analysis of coenzyme Q redox status
We used sheep heart mitochondria because they contain relatively large amounts of CoQ10, which expedited analysis. For evaluation of CoQ10 redox ratios, frozen mitochondria were thawed at 37°C and diluted with 500 µl buffer A to create a mitochondrial stock solution, which was kept on ice until use. For reactions, 50 µl of this stock suspension was added to 500 µl of buffer A, containing 0.5 µg/ml antimycin A from a 25 µg/ml stock solution in ethanol. Antimycin A binds to the Qi site of cytochrome c reductase (complex III), thereby inhibiting the upstream oxidation of any produced ubiquinol. Pa was added (25 µM final concentration) from a 1.3 mg/ml stock solution in DMSO. The suspension was added to a test tube, mixtures purged with argon and the reactions initiated by placing the tube between two LED light bulbs (previously described). We irradiated the samples for 10 minutes at room temperature. For negative controls, we repeated the above sequence changing the following: (1) in the absence of light; (2) in the absence of added P-a; (3) with heat denatured mitochondria; and (4) in the absence of added mitochondria but with added coenzyme Q. For a positive control we added 10 µl of a stock solution of 0.25 M glutamate/0.125 M malate in Tris buffer at pH 7. For mitochondrial denaturing, 200 µl of stock mitochondrial suspension was purged with argon and placed in a bath at 70°C for 5 minutes. For control reactions without added mitochondria, a coenzyme Q stock solution in buffer A was prepared by adding ALL-QTM (DSM Nutritinal products, Switzerland), a water-soluble coenzyme Q solution containing 10% coenzyme Q, modified food starch, sucrose and medium chain triglycerides, to buffer A. For these reactions 50 µl of the water-soluble Coenzyme Q stock or the denatured suspension was used as above in place of the mitochondrial stock solution. All reactions were adjusted to give the same amount of coenzyme Q in the reaction mixture as measured by HPLC.
To quantify relative ubiquinone and ubiquinol concentrations, a 50 µl aliquot was taken from the reaction mixture and was added to 200 µl of 0.4 M perchloric acid and 100 µl isopropyl ether containing 1 mg of butylated hydroxytoluene/ml as an antioxidant. The solution was vortexed for 1 minute, centrifuged for 2 minutes at 15,000 r.p.m. and the organic phase analyzed by HPLC. HPLC conditions have been reported previously (Qu et al., 2009; Qu et al., 2011). Briefly, we used an isocratic elutent consisting of 1% sodium acetate 3% glacial acetic acid, 5% butanol in methanol at 0.6 ml/minute. The HPLC column was 50×2.1 mm, C-18, 2.6 u, 100 Å (Phenomenex, Torrance, CA). A PDA detector set at 290 nm for ubiquinol and 275 nm for ubiquinone was used. We determined relative ubiquinol and ubiquinone concentrations by their online absorption spectra using extinction coefficients of 14,200 M−1 cm−1 at 275 nm in ethanol for ubiquinone and 4640 M−1 cm–1 at 290 nm in ethanol for ubiquinol (Lester et al., 1959).
Analysis of ATP synthesis in mouse brain homogenates
To produce homogenates of mouse brain, the frontal lobe was homogenized using two strokes of a Potter S homogenizer (Sartorius AG, Goettingen, Germany) at 4°C (20 mg of brain to 1 ml buffer A). The homogenate (80 µl) was added to buffer A (920 µl) and treated as described above for liver samples. Reactions were run in triplicate and data obtained between 5 and 50 minutes after lysis. ATP production showed a linear increase during this time, which was fitted to a line, the slope of which is reported as the relative ATP synthesis rate.
Analysis of ATP synthesis in mouse lens and heart homogenates
Lenses from mice were homogenized (KONTES® DUALL® tissue grinder with glass pestle) in ATP assay buffer (0.15 mM sucrose, 0.5 mM EDTA, 5 mM magnesium chloride, 7.5 mM sodium phosphate, 2 mM HEPES) at 50 µl buffer per lens. We added 1 µl of P-a stock (1 mM) and 1 µl of ADP stock (10 mM) to 100 µl lens homogenate. The mixture was exposed to red light (671 nm at 0.8 W/m2) or kept in dark for 20 minutes. ATP concentrations were determined using a luciferase-based ATP quantification kit according to the manufacture’s instructions (Life Technologies, Grand Island, NY).
Heart tissue (20 mg) was homogenized as above in 1 ml ATP assay buffer. 10 µl of P-a (1 mM) and 10 µl of ADP (10 mM) and 940 µl of ATP assay buffer were added into 40 µl tissue homogenate. The mixture was exposed to red light and ATP was determined as described above using a luciferase based ATP kit.
Analysis of ATP concentrations in duck adipose
We removed visceral fat from a duck (Anas platyrhynchos domestica) less then 30 minutes after death by decapitation and homogenized the fat at 4°C (without buffer) in a loose-fitting Potter-Elvehjem homogenizer. We then added P-a (70 µl of a 3.3 mg/ml stock solution) and ADP (800 µl of a 10 mg/ml stock solution). The homogenate was divided into two groups: one group was kept in the dark, while the other was exposed to red light (671 nm at 0.8 W/m2); both dishes were kept at 37°C. 200-µl aliquots were taken from each dish and ATP was measured using the luciferase assay or by HPLC, as described in the literature (Ally and Park, 1992).
Analysis of the effect of light wavelength
The entire brain of a mouse was homogenized with a Dounce homogenizer (20 mg of brain to 1 ml buffer C: 0.15 mM sucrose, 0.5 mM EDTA, 5 mM MgCl2, 7.5 mM Na2HPO4, 2 mM HEPES) at 4°C. We took a 40-µl aliquot of the homogenate and added it to 940 µl buffer C. We added 10 µl P-a (from a 1 mM stock in DMSO) and placed the sample on ice for 1 hour. We then added 10 µl ADP (from a 10 mM stock). Five 100-µl portions of the suspension were added to each well of a 96-well plate and exposed to light for 40 minutes. Then, 20-µl aliquots of the mixture were lysed with 200-µl lysis buffer for 1 hour on ice, and ATP levels were determined as above using a luciferase-based ATP kit.
Analysis of red fluorescence in tissue extracts
The chlorophyll-rich diet (Harlan Teklad, Indianapolis, IN) contained 15% by weight spirulina [a food supplement produced from cyanobacteria (Ciferri, 1983)], which is equivalent to ∼0.15% by weight chlorophyll-a. The control diet was a purified diet devoid of dietary chlorophylls (Harlan Teklad). The swine chlorophyll-rich diet has been described previously (Mihai et al., 2013).
For fluorescence spectroscopy, five pigs each were given these respective diets ad libitum for 2 weeks. Whole brain or 2–7 grams of abdominal fat was homogenized with a hand-held homogenizer (Omni Micro Homogenizer (μH), Omni International, Kennesaw, GA), HPLC grade acetone (40 ml) was added and the sample was vortexed for 1 minute. Insoluble material was precipitated by centrifugation and the acetone evaporated with a rotary evaporator. The samples were resuspended in 3 ml chloroform and measured directly.
For HPLC and UV spectroscopy, we extracted 2.5 grams of fat, as described above, from rats or swine that had been given a chlorophyll-rich diet, to give a clear oil. We then added 10 ml of absolute ethanol, cooled the sample to −20°C for 30 minutes, pelleted the insoluble material by centrifugation, separated and evaporated the ethanol with a rotary evaporator and re-suspended the sample in 500 µl of absolute ethanol. For plasma, we added 4 ml of plasma to 1 ml of saturated NaCl and 10 ml ethyl acetate, vortexed the sample for 1 minute and separated the layers by centrifugation. We removed the ethyl acetate layer, evaporated the ethyl acetate and re-suspended the resulting film in 300 µl of absolute ethanol. The samples were then used for HPLC and UV spectroscopy. A Waters (Milford, MA) HPLC system with a 600 pump, a 2475 fluorescent detector, a 2998 photodiode array (PDA) detector and a C18, 2.6 u, 100 Å, 150×2.10 mm column (Phenomenex, Torrance, CA) was used for HPLC. Excitation was set to 410 nm and emission set to 675 nm. Absorbance between 275 and 700 was recorded. We used a mobile phase of acetonitrile containing 10% isopropyl alcohol and 0.1% formic acid (solvent A) and water containing 0.1% formic acid (solvent B). Compounds were eluted at a flow rate of 0.3 ml/minute with a 50∶50 mixture of A∶B for 5 minutes, which was changed linearly to 100∶0, A∶B over 15 minutes. At 35 minutes, the flow was increased to 0.5 ml/minute.
In vivo imaging
Animals were imaged with a Maestro™ In-Vivo Imaging System (CRi, Hopkinton, MA), as described by Bouchard et al.; the animals were skinned to reduce interference from skin autofluoresence (Bouchard et al., 2007).
General C. elegans maintenance
Worms were a gift from Dr Cristina Lagido (Department of Molecular and Cell Biology, University of Aberdeen Institute of Medical Sciences, Foresterhill, Aberdeen, UK) (Lagido et al., 2009; Lagido et al., 2001). Nematode husbandry has been described previously (Wood, 1988). Briefly, animals were maintained on nematode growth medium (NGM) agar (Nunc) using E. coli strain OP50 as a food source. To obtain synchronous populations, we expanded a mixed population on egg yolk plates (Krause, 1995). Worm eggs were isolated from the population by treatment with 1% NaOCl/0.5 M NaOH solution (Emmons et al., 1979) and transferred to a liquid culture with E. coli strain OP50, carbenicillin (50 µg/ml) and amphotericin B (0.1 µg/ml; complete medium).
Real-time ATP monitoring in C. elegans
We administered the P-a chlorophyll metabolite by adding it to the culture medium for a minimum of 24 hours. To confirm P-a uptake, we washed away the culture medium containing P-a, suspended the worms in fresh medium and determined the fluorescence spectra in the worms. Treated worms had signature chlorophyll-derived fluorescence, whereas control worms that were not given P-a exhibited no such fluorescence, confirming metabolite uptake.
Method A
Worms were grown in liquid culture at a density of 10,000 worms/ml. Twenty-four hours before the experiment, the culture was split into control and treatment groups and varying amounts of a P-a stock solution in DMSO were added to the treated groups. Control worms were given DMSO vehicle. Worms were washed with M9 buffer (IPM Scientific, Eldersburg, MD) to remove food and unabsorbed P-a and resuspended at 3000 worms/ml. 50 µl of worm suspension from each of these groups were plated into a well of a 96-well plate. Each experimental group was plated into a minimum of 12 wells. To assay ATP stores by luminescence, 100 µl of luminescence buffer containing D-luciferin was added to each well, according to the literature (Lagido et al., 2009; Lagido et al., 2001) and luminescence was recorded in a plate reader. The luminescence buffer was a citric phosphate buffer at pH 6.5, 1% DMSO, 0.05% Triton X-100 and D-luciferin (100 µM). After initial ATP measurements, half of the worms from each experimental group were exposed to LED light centered at 660 nm at 1±2 W/m2; the other half was kept in the dark by covering the plate with aluminium foil. ATP (luminescence signal) was recorded periodically. The amount of ATP synthesized was reported as the difference within an experimental group between the luminescence signal of worms kept in the dark and the worms exposed to light. All experimental procedures outside of red light exposure were performed under dim light. The experiment was repeated three times with different populations of worms.
Method B
Worms were plated as above, with each experimental group divided into 12 wells of a 96-well plate. Four identical 96-well plates were made, each containing worms treated with varying concentrations of P-a and control worms. At time zero, 100 µl of luminescence buffer was added to a plate and in vivo ATP was assayed as luminescence. The remaining three plates were exposed to light and ATP assays were performed every 15 minutes for 45 minutes by the addition of 100 µl of luminescence buffer and the recording of luminescence.
In vitro ATP monitoring in C. elegans
One-day-old adult worms in liquid culture were incubated with P-a for 24 hours, washed with M9 buffer and re-suspended in M9 buffer at 50,000 worms/ml. The control group was incubated in DMSO vehicle without P-a. 100 µl of each worm suspension was placed into 18 centrifuge tubes. At time zero, six tubes from each group were placed in liquid nitrogen and the remaining tubes exposed to red light. Then, at 15 and 30 minutes, six tubes from each group were placed into liquid nitrogen. To measure ATP, we removed the centrifuge tubes from the liquid nitrogen and placed them in boiling water for 15 minutes to lyse the worms (Artal-Sanz and Tavernarakis, 2009). The resulting solution was cleared by centrifugation for 5 minutes at 15,000 rpm and ATP in the lysate was measured using the luciferase assay according to the manufacturer’s instructions or by HPCL according to established protocols (Ally and Park, 1992).
Analysis of C. elegans oxygen consumption
Oxygen consumption was measured using a Clark-type oxygen electrode (Qubit Systems Inc.), as described (Anderson and Dusenbery, 1977; Zarse et al., 2007). One-day-old adult worms in liquid culture at a density of ∼10,000 worms/ml were incubated with P-a (25 µM) for 24 hours in complete medium. Animals were washed three times with M9 buffer to remove bacteria and excess P-a and resuspended in M9 buffer at 10,000 worms/ml. One-ml aliquots of this suspension were transferred into the respiration chamber and respiration was measured at 25°C for 10 minutes while being exposed to an LED light centered at 660 nm at 1±2 W/m2. The control group was treated in the same way but not incubated with P-a.
Analysis of ROS formation in C. elegans
ROS formation was quantified as described by Schulz et al. (Schulz et al., 2007). Three-day-old worms were synchronized in liquid culture at a density of 500 worms/ml in complete medium, then divided into control and treatments groups. The treatment group was incubated for 24 hours with 12 µM P-a and the control group in DMSO vehicle. Bacterial food and P-a were removed by three repeated washes with M9 and the worms resuspended to 500 worms/ml M9 buffer. 50 µl of the suspension from each group was added to the wells of a 96-well plate with opaque walls and a transparent bottom. A 100 µM 2′,7′-dichlorofluorescin diacetate (Sigma-Aldrich, St. Louis, MO) solution in M9 buffer was prepared from a 100 mM 2′,7′-dichlorofluorescin diacetate stock solution in DMSO. 50 µl of this solution were pipetted into the suspensions, resulting in a final concentration of 50 µM. Additional controls included worms without 2′,7′-dichlorofluorescin diacetate and wells containing 2′,7′-dichlorofluorescin diacetate without animals; these were prepared in parallel. Five replicates were measured for each experimental and control group. Immediately after addition of 2′,7′-dichlorofluorescin diacetate, the fluorescence was measured in a SpectraMax M5 microplate reader (Molecular Devices, LLC, Sunnyvale, CA) at excitation and emission wavelengths of 502 and 523 nm. The plates were then exposed to red LED light and fluorescence was re-measured at 2.5 and 5 hours under conditions equivalent to those used previously.
Life span analysis
Population studies
Life span measurements were performed according to the method of Gandhi et al. and Mitchell et al. (Gandhi et al., 1980; Mitchell et al., 1979) with some modifications. Eggs were harvested and grown in darkness in a liquid culture at room temperature. To prevent any progeny developing, 5-fluoro-2′-deoxyuridine (FUDR) (Sigma-Aldrich, 120 µM final) was added at 35 hours after egg isolation, during the fourth larval molt. At day 4 of adulthood, the culture was split into control and experimental groups. The experimental group was treated with 12 µM P-a from a stock solution in DMSO. The control group was given the DMSO vehicle alone. The treated and control cultures were then split into two or three. The final density of worms in all reaction flasks was 500 worms/ml; each flask contained 10 ml, therefore a total of 5000 worms. The following day (day 5 of adulthood), worms were exposed to LED light centered at 660 nm at 1±2 W/m2 for 5 hours. Light exposure was repeated every day until the end of the experiment. For counting, aliquots were withdrawn and placed in a 96-well plate to give ∼10 worms per well; the worms were scored dead or alive on the basis of their movement, determined with the aid of a light microscope. A total of 60–100 worms (representing 1–2% of the total population) were withdrawn and counted at each time point for each flask. Counts were made at 2–3-day intervals and deaths were assumed to have occurred at the midpoint of the interval. Any larvae that hatched from eggs produced before the FUDR was added remained small in the presence of FUDR and were not counted. We used the L4 molt as time zero for life span analysis. To obtain the half-life, we plotted the fraction alive at each count verses time and fitted the data to a two-parameter logistic function using the software GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). The two-parameter model is known to fit survival of 95% of the population fairly accurately (Vanfleteren et al., 1998). Because changes in environment, such as temperature, worm density and the amount of food, can influence life span, control measurements were conducted at the same time under identical conditions. The concentration of P-a dropped (∼75%) throughout the life span studies and it was not adjusted (supplementary material Fig. S4F).
Life span measurements in 96-well microtiter plates
Life span was measured as described in the literature (Solis and Petrascheck, 2011), except that P-a was added at day 4 and light treatment commenced at day 5. Scoring (fraction alive) was done once on day 15.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
C.X. conducted studies with worms, and ATP measurements in mitochondria and tissue homogenates. J.Z. conducted ATP measurements in mitochondria and tissue homogenates. D.M. conducted metabolite-binding distribution studies. I.W. designed and supervised the study and wrote the manuscript.
Funding
This work was supported by the Department of the Navy, Office of Naval Research [grant number N00014-08-1-0150 to I.W.]; the Nanoscale Science and Engineering Initiative of the National Science Foundation [grant numbers CHE-0117752, CHE-0641532 to I.W.]; and the New York State Office of Science, Technology and Academic Research (NYSTAR).
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.134262/-/DC1
- Received April 30, 2013.
- Accepted October 15, 2013.
- © 2014. Published by The Company of Biologists Ltd
References
Sunlight is the most abundant energy source on this planet. However, the ability to convert sunlight into biological energy in the form of adenosine-5′-triphosphate (ATP) is thought to be limited to chlorophyll-containing chloroplasts in photosynthetic organisms. Here we show that mammalian mitochondria can also capture light and synthesize ATP when mixed with a light-capturing metabolite of chlorophyll. The same metabolite fed to the worm Caenorhabditis elegans leads to increase in ATP synthesis upon light exposure, along with an increase in life span. We further demonstrate the same potential to convert light into energy exists in mammals, as chlorophyll metabolites accumulate in mice, rats and swine when fed a chlorophyll-rich diet. Results suggest chlorophyll type molecules modulate mitochondrial ATP by catalyzing the reduction of coenzyme Q, a slow step in mitochondrial ATP synthesis. We propose that through consumption of plant chlorophyll pigments, animals, too, are able to derive energy directly from sunlight.